The structural characterization of the cellular machinery is fundamental to understand its mechanism of action. X-ray crystallography, NMR and more recently EM have revolutionised the field of structural biology. However, all these methods require the isolation of the protein complex. Thus, the structures of the cellular machinery are often determined in an environment that does not resembles the crowded interior of a cell where those assemblies are actually functional.
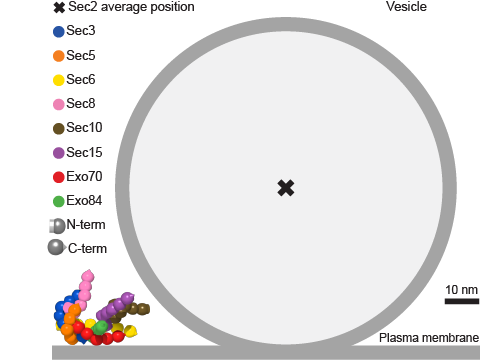
In collaboration with the group of Marko Kaksonen and Damien P. Devos, we have developed a new approach to determine the architecture of multisubunit complexes directly in living cells. As a proof-of-principle we have determined the molecular architecture of the COG (conserved oligomeric Golgi) complex for seven of its eight subunits (Figure 1). We have also determined the architecture of the exocyst bound to a vesicle (Figure 2).
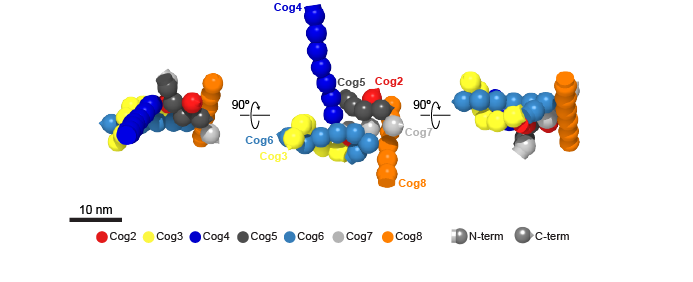
We continue working on the development of this approach in order to increase the precision of the structural reconstructions determined in vivo. In this direction, we aim to integrate our approach with other techniques such as crystallography, EM and super-resolution microscopy to reach a complete mechanical understanding of those processes that mediate cellular compartmentalization.