Since specific recognition between proteins governs cellular function, the systematic study of protein-protein interactions has become a central endeavor in the fields of structural and cell biology. However, although the crowded cellular environment is expected to play a major role in protein function, protein complexes are usually characterized using in vitro methods with isolated proteins. In addition researchers frequently fail to isolate and characterize short-lived complexes that remain as one of the major bottlenecks for modern structural biology.
To overcome aforementioned limitations we use PICT (Protein interactions from Imaging of Complexes after Translocation), a light microscopy method that allows quantitative measurements of transient assemblies in living cells (Gallego et al. 2013). PICT takes advantage of the inducible dimerization of FK506-binding protein (FKBP) and FKBP-rapamycin binding (FRB) domain to translocate protein assemblies to membrane associated anchoring platforms (Figure 1).
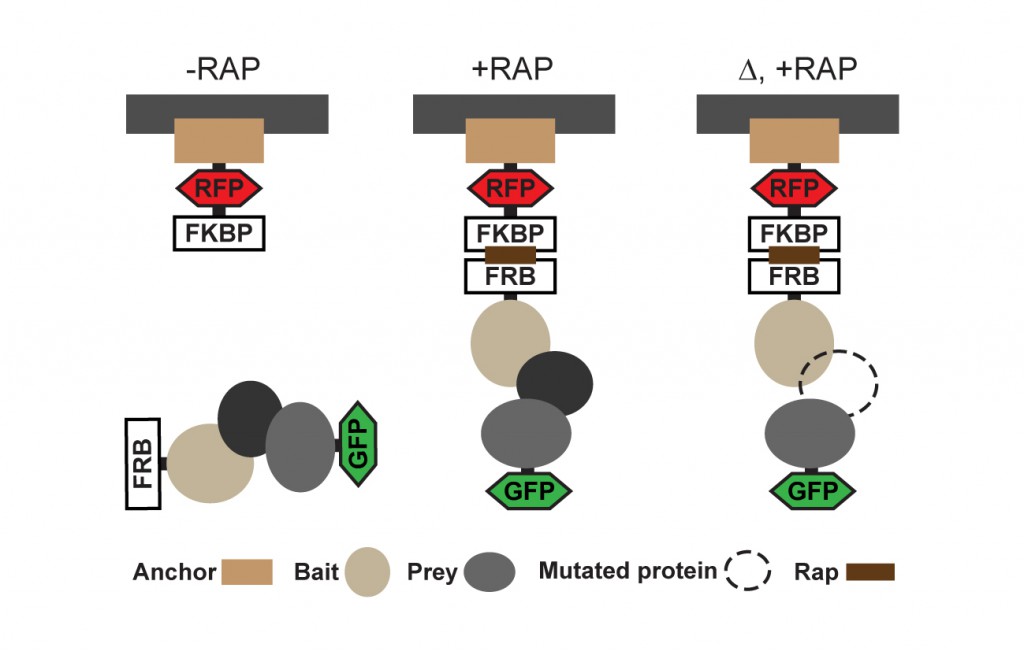
In this assay, GFP- tagged prey proteins interacting with the FRB–tagged bait co-translocate to the FKBP-tagged anchor sites upon addition of rapamycin. The interactions are thus encoded into localization changes and can be detected by fluorescence live-cell imaging under physiological conditions or upon perturbations (Movie 1, adapted from Gallego et al. 2013).
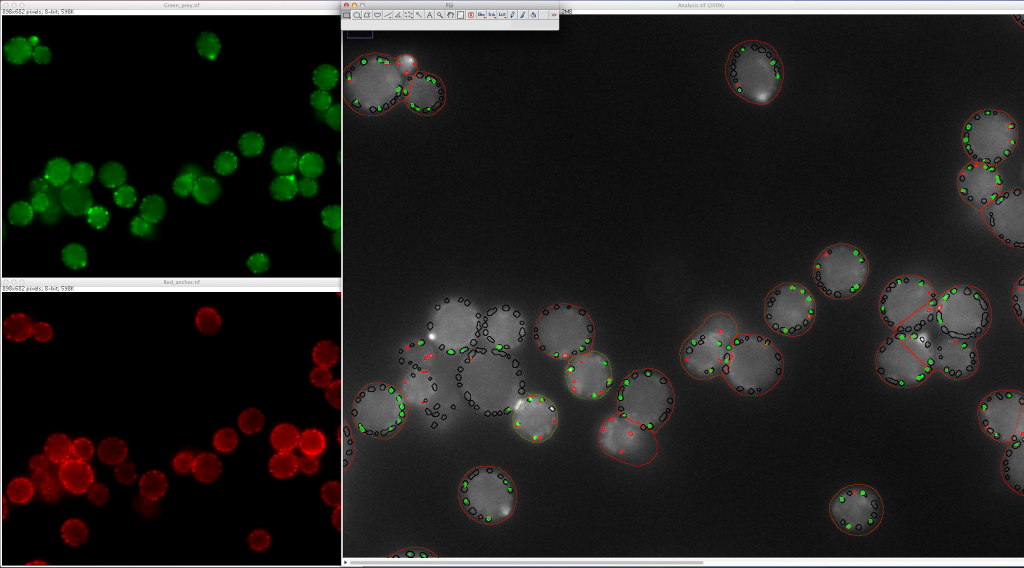
PICT provides a unique combination of advantages: PICT is based on simple, fast and noninvasive fluorescence microscopy readout. The simplicity of the readout also makes automation of PICT for high-throughput studies straightforward (Figure 2). Perhaps most importantly, the method can provide quantitative measurement such as dissociation rates under native conditions.